Abstract
Background: Next-generation sequencing (NGS) is an attractive tool for prospective use in the field of precision medicine. Using NGS to guide therapy has provided a large volume of genomic data and therapeutic actionability of somatic NGS results. These data are evolving too rapidly to rely solely on human curation. So the interpretation of the clinical significance of such large amounts of genetic data remains the most severe bottleneck preventing the realization of precision medicine. Watson for Genomics (WfG) is a representative artificial intelligence (AI) software, which analyzes and categorizes genetic alterations that are related to disease progression and provides a list of potential therapeutic options within 3 minutes per sample. Recent reports suggested that WfG could empower tumor boards and improve patient care by providing a rapid, comprehensive approach for data analysis and consideration of the up-to-date availability of clinical trials (Patel NM, et.al. Oncologist. 2018). However, only limited data are available regarding the utility of AI-guided precision medicine approach in the field of hematological disease. The purpose of this study is to test the utility of AI in assisting the interpretation of high throughput genomic data from patients with the hematological disease.
Methods: After obtaining written informed consent, we enrolled patients with hematological disease at our research hospital between May 2015 to June 2018. Genomic DNA was prepared from malignant cell fractions and normal tissues in each patient and subjected to comparative NGS, mainly targeted deep sequencing (TDS) with ready-made panels and, on demand, whole exome sequencing (WES). Sequence data was analyzed using a pipeline of in-house semi-automated medical informatics. After initial bioinformatics filtering, we used WfG to identify potential driver mutations, which were annotated as "pathogenic" or "likely pathogenic" (WfG version 39.132 and 39.135 as of July 2018). The results were compared with the findings of expert hematologists.
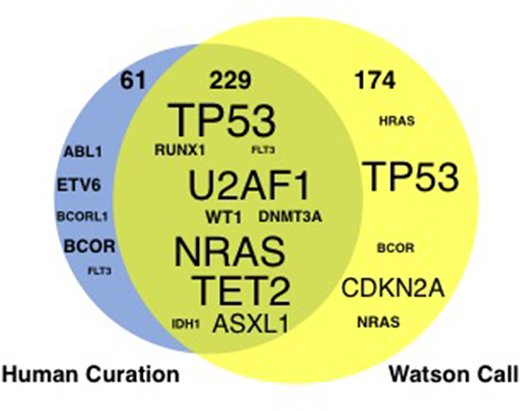
Results: 247 paired samples (TDS, n= 143; WES, n= 104) collected from 187 patients were analyzed. Our cohort consisted of 63 patients with acute myeloid leukemia, 40 with myelodysplastic syndromes (MDS), 19 with myeloproliferative neoplasms (MPN), 9 with MDS/MPN, 10 with acute lymphoblastic leukemia/lymphoma, 17 with non Hodgkin lymphoma, 6 with multiple myeloma (MM) and others. In 151 of 187 patients, a total of 290 somatic driver mutations were identified by human curation. The frequently mutated genes were TP53 (n=31), NRAS (n=17), TET2 (n=16), U2AF1 (n=14), FLT3/ASXL1/WT1 (n=13 each), and DNMT3A/RUNX1 (n=12 each). WfG identified 79% (n=229) of driver mutations which human experts also did. There was some discordance between WfG and the human (Figure 1): Sixteen mutations were interpreted as "variant of unknown significance" by WfG, but these mutations were deduced as driver mutation by the human. Conversely, in two representative cases, WfG identified a relevant driver mutation that the human did not: FAM46C and SOCS1, from a patient with MM and with primary mediastinal large-B cell lymphoma, respectively. These examples indicate the potential for a mutually complementary or cooperative relationship between AI "software" and the human expert "hardware" in the interpretation of high throughput genomic data.
Conclusion: Combing AI "software" and the human expert "hardware" will allow for the quick delivery of comprehensive information needed for patient care that outperforms what either can achieve individually in the field of hematological disease.
Figure1. Comparison of potential driver mutations between human curation and Watson for Genomics. The size of the gene symbol indicates the total number of mutations identified
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.